Time:2017-04-07
A recent study by Dr. YAN Jun’s lab at the Institute of Neuroscience, Chinese Academy of Sciences, was published in Nucleic Acids Research on March 8. It uncovered that a class of enhancer-associated circadian long noncoding RNAs (lncRNAs) mark the genomic loci modulating long-range circadian gene regulation.
Circadian rhythm is an intrinsic 24h oscillation of various physiological processes and behaviors synchronized with daily light/dark cycle. It exerts its influence by regulating gene expression at various levels. lncRNAs are defined as RNA transcripts longer than 200 bp but lacking protein coding potential. They are expressed highly tissue-specifically and have been found playing roles in many biological process such genome printing, cell cycle and tumor genesis. Though several specific circadian lncRNAs have been found, there is no systematic study on lncRNAs with circadian expression, as previous study in circadian regulation are more focused on protein coding genes. In this study researchers systematically examined the expression of circadian lncRNAs in mouse liver and investigated role in regulating circadian rhythm.
After sequencing the transcriptome of mouse liver, researchers systematically assembled lncRNAs and found those with oscillation in 24 hours. Many of the circadian lncRNAs were bound by two key circadian transcription factors, BMAL1 and REV-ERBα. By analyzing enhancers defined by histone marks, a significant proportion of circadian lncRNAs were found expressed at enhancer regions. Though tissue specificity of lncRNAs and enhancers, this association between circadian lncRNAs and enhancers can also be observed in other tissues. Circadian lncRNAs showed strong correlation with nearby genes and showed similar circadian phases with them. lncRNAs were found showing higher extent of nuclear localization than protein coding genes, exemplified in Figure A. By comparing lncRNAs in mouse and rat, researchers found that lncRNAs transcription loci tended to be conserved though lncRNAs sequences were of poor conservation. Researchers identified one such candidate circadian lncRNA named lnc-Crot that was transcribed from a super-enhancer region. By 4C-seq (Figure B) this region was found interacting with genes related with energy metabolism through long-range interactions. And the interacted genes tended to show similar circadian phase with lnc-Crot. Further experiments in cell line showed this interaction facilitate functions of circadian transcription factors that bind on the lnc-Crot region. This study systematically explored circadian lncRNAs in mouse liver and identified a group of them mark active enhancers that are involved in circadian regulation through long-range interactions. Though these interactions are relatively stable, enhancers could facilitate the function of transcription factors, which were bound on on the enhancers, to confer time variant regulation (Figure C).
This study has summarized and published on NAR journal entitled with “A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation”, was mainly carried out by FAN Zenghua, ZHAO Meng and other members under the supervision of Dr. YAN Jun in collaboration with the group led by Dr. ZHAO Zhihu at the Academy of Military Medical Sciences and group led by Dr. Gregor Eichele at Max Planck Institute for Biophysical Chemistry. And the work was supported by the grant from Chinese Academy of Sciences Strategic Priority Research Program and Natural Science Foundation of China Grant.
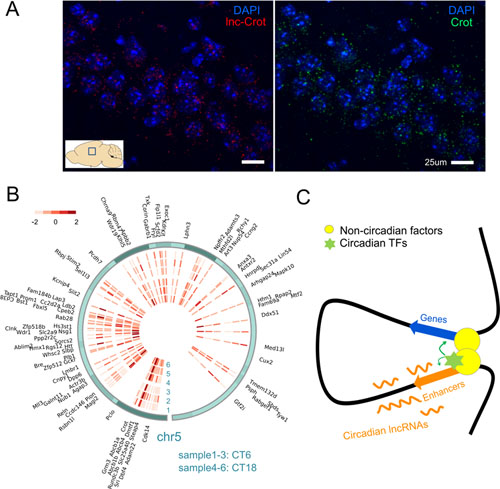
(A) Sub-cellular expression of lnc-Crot and Crot by FISH(Fluorescence In Situ Hybridization). (B) Heatmap of interaction intensities of 4C-seq and interacted genes on chr5. (C) One hypothetical model that enhancers marked by circadian lncRNAs modulate circadian gene expression through long-range interaction by facilitating functions of circadian transcription factors bound on them.
 附件下载:
附件下载: