Time:2020-03-14
A recent study published in PLOS Biology revealed that light-dark cycle is essential for the initiation of the cellular clock in zebrafish by constructing a transgenic zebrafish line to report in vivo circadian rhythm at the single-cell level. This work was performed by researchers in Dr. YAN Jun’s Lab and Dr. HE Jie’s lab at the Center for Excellence in Brain Science and Intelligence Technology, Institute of Neuroscience, State Key Laboratory of Neuroscience, Chinese Academy of Sciences in collaboration with Dr. LI Yuanhai at First Affiliated Hospital of AnHui Medical University.
Circadian rhythm evolves to align animal behaviors to periodic daily environmental changes. At the molecular level, vertebrate circadian clock is mainly generated through transcriptional/translational feedback loops of core clock genes. At the organismal level, the overall circadian rhythm is governed by an intricate network of circadian oscillators, in which the master pacemaker such as SCN in mammals or the pineal gland in zebrafish is believed to play a pivotal role. It has been shown that circadian oscillation was established gradually during development, however how single-cell circadian clocks were established during embryonic development is still unclear. Zebrafish (Danio rerio) embryos that are in vitro fertilized and transparent provide an accessible model organism for in vivo live imaging, and thereby being widely used in the study of animal development. It is well-documented that early exposure to light-dark (LD) cycle is required for the development of circadian clock in zebrafish larva. But it is still under debate whether the effect of light on clock development is a synchronization of already existing oscillators or an initiation of single-cell clocks. Questions like these can only be addressed by time-lapse in vivo imaging of single-cell circadian clock reporter.
In the present study, they report a transgenic zebrafish line using destabilized fluorescent protein, Venus-NLS-PEST (VNP), driven by the promoter of a key circadian clock gene, nr1d1 (Figure A). They observed that VNP reporter expression undergoes stepwise onset starting at the photoreceptor cells in the pineal gland then spread to cells in other brain regions (Figure B,C). By averaging 3D cell density distribution of nr1d1:VNP-positive cells across six fish, they found that nr1d1:VNP-positive cells were mostly distributed around the lumen of the pineal gland while the cell density was relatively low in the central part of the pineal gland (Figure D). Using single-cell RNA-seq, they characterized the cell types expressing VNP in the whole brain (Figure E). Within the pineal gland, they found that each cell undergoes circadian oscillation superimposed over cell-type specific developmental trajectories (Figure F). Under light-dark (LD) cycle, cellular expression of VNP shows synchronous circadian oscillation. However, the circadian expression of nr1d1:VNP-positive cells were dramatically dampened rather than desynchronized while the developmental trend was still present at single cell level in fish raised under constant darkness (DD) (Figure G). Their result suggests that the early exposure of LD cycle is crucial for the ontogeny of functional single-cell oscillators.
In this work, they constructed live zebrafish carrying nr1d1:VNP fluorescence reporter, which allows for monitoring the circadian gene expression at the single-cell resolution in zebrafish larvae for the first time. Using this reporter fish, they revealed the interplays among circadian clock, cell-type specific development, and light-dark cycle. This newly developed nr1d1:VNP fish can be used to examine the relationship between circadian clock and cell cycle in vivo, conduct cell-type specific imaging of circadian rhythm, and screen for chemical compounds affecting circadian rhythm and sleep. In summary, their single-cell circadian reporter zebrafish line will have broad applications in circadian research.
This work entitled “Single-cell in vivo imaging of cellular circadian oscillators in zebrafish” was published online in PLOS Biology on March 14, 2020. WANG Haifang, Dr. YANG Zeyong and LI Xingxing are the first authors with equal contribution under the supervision of Dr. YAN Jun, Dr. HE Jie, and Dr. LI Yuanhai. HUANG Dengfeng and YU Shuguang also contributed to this work.
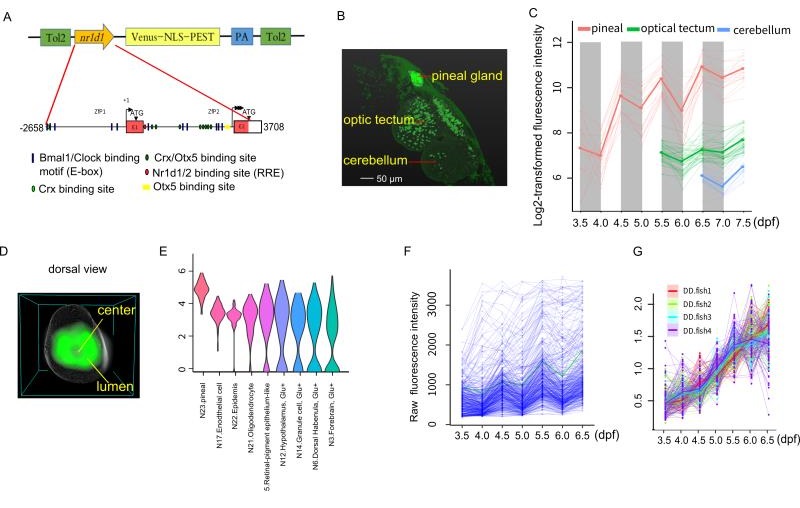
Figure legend: (A) The schematic of nr1d1:VNP construct design. (B) Whole-brain two-photon image shows the spatial distribution of nr1d1:VNP-positive cells in different brain regions at 7.5dpf. (C) Single-cell tracing result of nr1d1:VNP-positive cells in the whole-brain. (D) 3D reconstruction of a common zebrafish pineal gland by aligning and averaging of six fish. The grey sphere represents the boundary of the pineal gland and the green color represents the density distribution of nr1d1:VNP-positive cells in the pineal gland. (E) Violin plot demonstrated the clusters with enriched expression of nr1d1:VNP. (F) Raw fluorescence intensity of all the traced nr1d1:VNP-positive cells cells under LD condition (six fish). (G) Expression patterns of all the traced nr1d1:VNP-positive cells under DD condition during development (four fish).
AUTHOR CONTACT:
YAN Jun
Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai, China.
E-mail: junyan@ion.ac.cn
 附件下载:
附件下载: